|
|
||
|
New species of bacteria found in Titanic 'rusticles' ..
New species of
bacteria found
in Titanic 'rusticles'
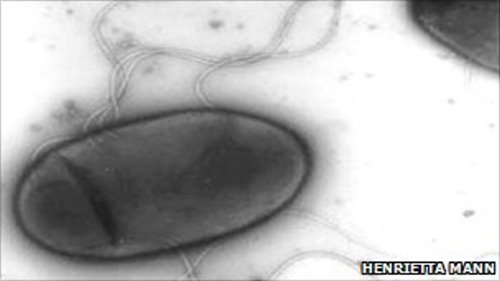
The Halomonas titanicae bacterium was found in "rusticles", the porous and delicate icicle-like structures that form on rusting iron. Various bacteria and fungi live within the delicate structures - first identified on the Titanic - actually feeding off the rusting metal. The find is described in the journal International Journal of Systematic and Evolutionary Microbiology. Samples of
rusticles from Titanic
were gathered in 1991 by the Mir 2 robotic
submersible.
Researchers from Dalhousie University and the Ontario Science Centre in Canada and the University of Seville in Spain isolated the H. titanicae bacteria from those samples. They sequenced the microbes' DNA before discovering that they constituted a new member of the salt-loving Halomonas genus. The bacteria are of particular interest because they may shed light on the mechanism by which rusticles form, and thus on the general "recycling" that such microbes carry out on submerged metal structures. That, the authors point out, has relevance also to the protection of offshore oil and gas pipelines, and the safe disposal at sea of ships and oil rigs. SOURCE: BBC News Related Links:
|
||
| Halomonas
titanicae sp. nov.,
a halophilic bacterium isolated from the RMS Titanic
Cristina Sánchez-Porro1, Bhavleen Kaur2, Henrietta Mann3 and Antonio Ventosa1,4 - 8 January 2010 1 University of Sevilla, Spain;
4 E-mail: ventosa@us.es A Gram-negative, heterotrophic, aerobic, non-endospore-forming and motile by peritrichous flagella bacterial strain, designated strain BH1T, was isolated from a rusticle sample, a consortium of microorganisms, collected from the RMS Titanic wreck site. The strain grew optimally at 30-37 °C, pH 7.0-7.5 and in the presence of 2-8% (w/v) NaCl. We carried out a polyphasic taxonomic study in order to characterize it in detail. Phylogenetic analyses based on the 16S rRNA gene sequence comparison indicated that strain BH1T clustered within the branch constituted by species of Halomonas. The most closely related species were Halomonas neptunia (98.6% 16S rRNA sequence similarity), Halomonas variabilis (98.4%), Halomonas boliviensis (98.3%) and Halomonas sulfidaeris (97.5%). The other closely related species were Halomonas alkaliphila (96.5% sequence similarity), Halomonas hydrothermalis (96.3%), Halomonas gomseomensis (96.3%), Halomonas venusta (96.3%) and Halomonas meridiana (96.2%). The predominant fatty acids of strain BH1T were C18:1 {omega}7c (36.3%), C16:0 (18.4%) and C19:0 cyclo {omega}8c (17.9%). The DNA G+C content was 60.0 mol%. Ubiquinone 9 (Q9) was the major lipoquinone. Phenotypic features, fatty acids profile and DNA G+C content further supported the placement of strain BH1T in the genus Halomonas. DNA-DNA hybridization between strain BH1T and the species Halomonas neptunia CECT 5815T, H. variabilis DSM 3051T, H. boliviensis DSM 15516T and H. sulfidaeris CECT 5817T were 19%, 17%, 30% and 29%, respectively, supporting its differential taxonomic status. On the basis of the phenotypic, chemotaxonomic, and phylogenetic features, a novel species is proposed to accommodate the strain, for which the name Halomonas titanicae sp. nov. is proposed. The type strain is BH1T (=ATCC BAA-1257T = CECT 7585T = JCM 16411T). SOURCE: http://ijs.sgmjournals.org/cgi/content/abstract/ijs.0.020628-0v2 |
||
| FAIR USE NOTICE: This page contains copyrighted material the use of which has not been specifically authorized by the copyright owner. Pegasus Research Consortium distributes this material without profit to those who have expressed a prior interest in receiving the included information for research and educational purposes. We believe this constitutes a fair use of any such copyrighted material as provided for in 17 U.S.C § 107. If you wish to use copyrighted material from this site for purposes of your own that go beyond fair use, you must obtain permission from the copyright owner. | ||
|
|